Overview
TopChem2
TopChem2 is a standalone program specifically designed to the quantum chemical topology analysis of the electron density, the electron localization function (ELF), the Molecular Electrostatic Potential (MESP) or the Reduced Density Gradient (NCI method). The Fukui functions and the Dual Descriptor are also computed in order to predict the reactivity of the reactants. The inputs files are the gaussian/Q-Chem wfn/wfx files link1 link2 or the three-dimensional cube files link3 link4. The program uses full algorithms enabling to assign the data points grids to basin volumes. It is distributed as binary only, free of charge. Easily usable in command-line, automatic and robust.
by J. Pilmé, Laboratoire de Chimie Théorique - Sorbonne Université.
Contact
email : julien.pilme@sorbonne-universite.fr
Try TopChem2 with TopChem Web
Input Files.....
Input files are the WFN/WFX files or the three-diminensional cube
files typically generated by several quantum chemistry codes :
- WFN/WFX Gaussian files: link
- WFN Q-Chem files: link .
- Gaussian cube files: link , use the utility cubegen for generating cubes files.
- Molden2AIM is designed to create AIM-WFN, AIM-WFX, and NBO-47 files from Molden files link
- NWChem cube files: link
- VASP: link ; CHGCAR and ELFCAR files can be directly used for generating cube files using the utility vasp_to_cube included in the TopChem2 package.
- CPMD cube files: link ; use the utility cpmd2cube for generating cubes files.
- Quantum Espresso: link
- ADF: link
- CP2K cube files: link
- Dirac cube files link
This list does not pretend to be exhaustive. ..
Output Files..
Several output files of TopChem2 can be processed by advanced free visualization code such as Molekel ,
VMD or VESTA programs. They can be used for rendering plots but they are not required to compute and produce chemical data. Critical points for the electron density and attractors for the electron localization function (included the degenerated attractors) are provided. Numerous local descriptors are computed at the critical points locations and are printed:
electron density, electron localization function, laplacian of the electron density, gradients and eigenvalues of the electron density and ELF/MEP, energy densities, ellipticity...
See Full list to --> Current Local Descriptors Implemented
The output file includes a Conceptual DFT (CDFT) section analysis. Some global CDFT descriptors are computed (chemical potential, softness..). The calculation of Fukui functions and the Dual Descriptor values are distributed on three dimensional grids.
TopChem2 is also interfaced with VMD. A VMD visualization state file called "file_elf_ebas.vmd" is automatically produced as an output file for the ELF/Dual Descriptor analysis. The color types will be automatically assigned and stored in five separated cube files. The vmd file can be directly read in VMD as a "Load visualization file" using ,
vmd -e file_elf_ebas.vmd
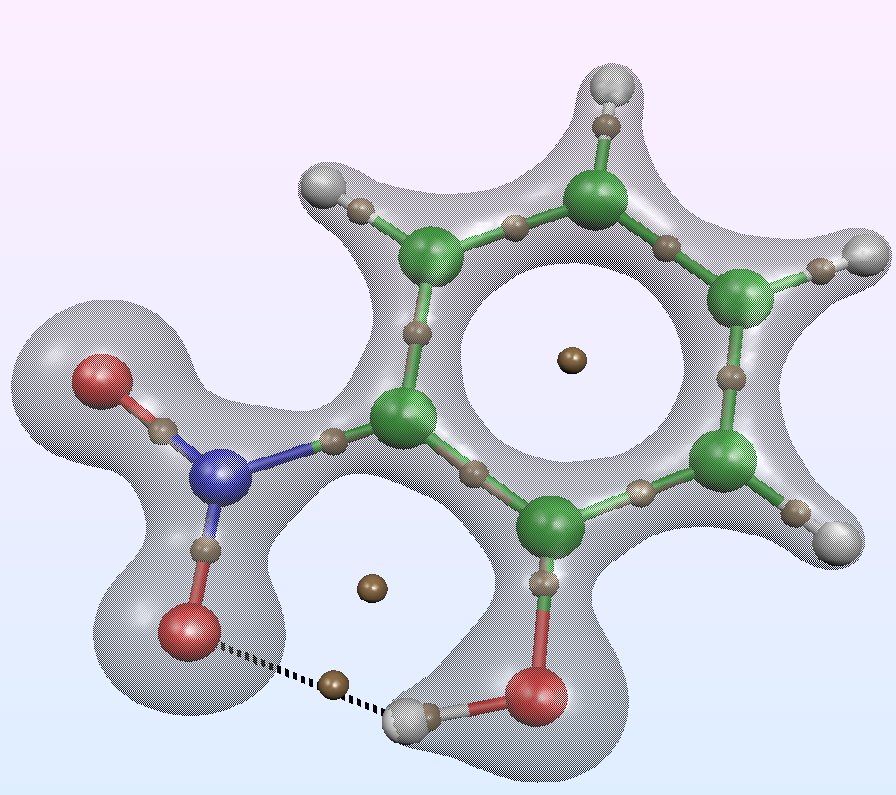
The Quantum Theory of Atoms in Molecules (QTAIM)
# Of all the possible ways to analysis the electron density in a molecule or in a solid, the Quantum Theory of Atoms in Molecules (QTAIM) is probably the most used for discussing the nature of chemical bonding . The pioneering works are due to Richard F. W. Bader and coworkers in the 70s. this methodology can be applied to both experimental and computed electron densities, the topological atoms being defined as the union of a nucleus and of its atomic basin. The topology of the density gradient field is characterized by critical points and their connectivity. Among the saddle points, a bond critical point (BCP) plays an essential role because the values of some local descriptors based on the electron density are usually related to the nature of the chemical bond.
See R. Bader, Chem. Rev. 91, pp. 893-928 (1991)
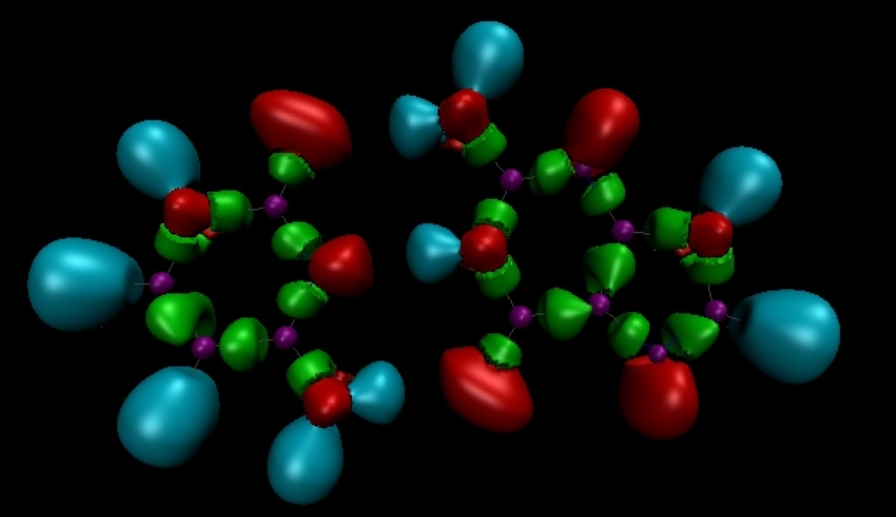
The Electron Localization Function (ELF)
# The electron localization function (ELF) topological analysis was intensively used for studying of the bonding schemes in molecules and in solids, or for rationalizing the chemical reactivity. It relies on the Laplacian of the conditional same spin pair probability scaled by the homogeneous electron gas kinetic energy density. ELF is usually interpreted as a signature of the electron-pair distribution. Its topological analysis represents a bridge between the traditional pictures of the chemical bond derived from the Lewis theory, and first principles quantum-mechanical methodologies. Overall, the spatial distribution of the valence basins closely matches the non-bonding and bonding domains of the VSEPR model.
See B. Silvi and A. Savin, Nature, 371, pp. 683–686 (1994).
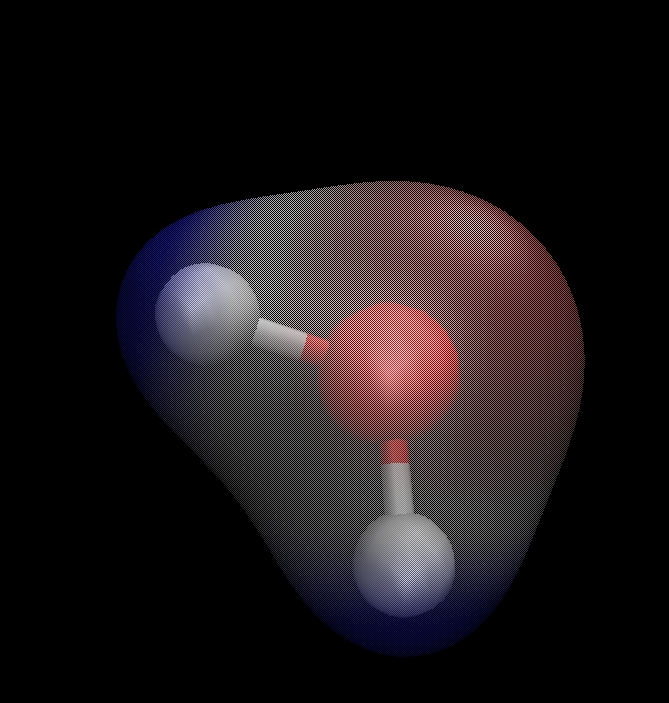
The Molecular Electrostatic Potential (MESP)
# The MESP is an interesting travel guide assessing the chemical reactivity of molecules towards positive (electrophilic) or negative (nucleophilic) sites of reactants. The MESP is typically visualized using mapping its values onto the electron density isosurface reflecting the molecular sites boundaries.
See H. Suresh, Geetha S. Remya, Puthannur K. Anjalikrishna, WIREs Comput Mol Sci 12:e1601 (2022)
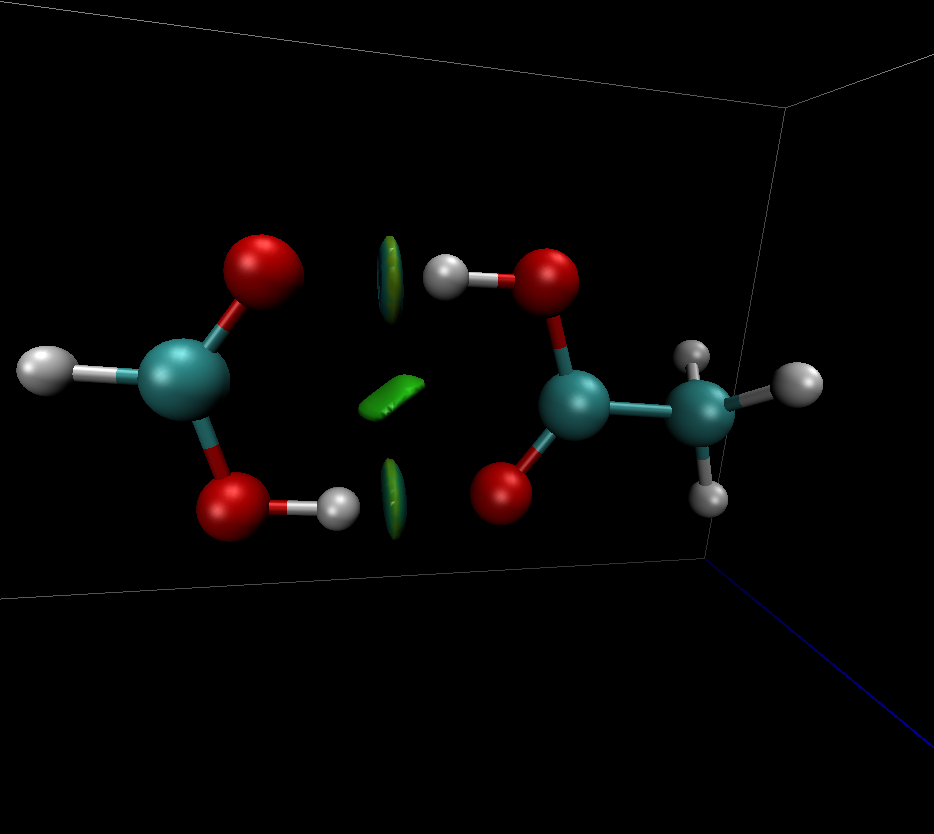
The Non-Covalent Interactions Index (NCI)
#NCI. The non-covalent interaction index (NCI) method relies of the properties of the reduced density gradient (RDG). RDG is very used for studying the weak interactions : The « strength » of the weak interactions can be visualized using a mapping of sign(λ2)*ρ(r) onto RDG isosurfaces, sign(λ2) being the sign of the second density Hessian eigenvalue (λ2). Typically, the attractive and repulsive interactions are identified as regions where λ2 < 0 and λ2 > 0, respectively. Weak van der Waals interactions when λ2 becomes nul.
See E. R. Johnson, S.R Keinan, P. Mori-Sánchez§, J. Contreras-García§, A. J. Cohen, and W. Yang J. Am. Chem. Soc. , 132, 6498 (2010).
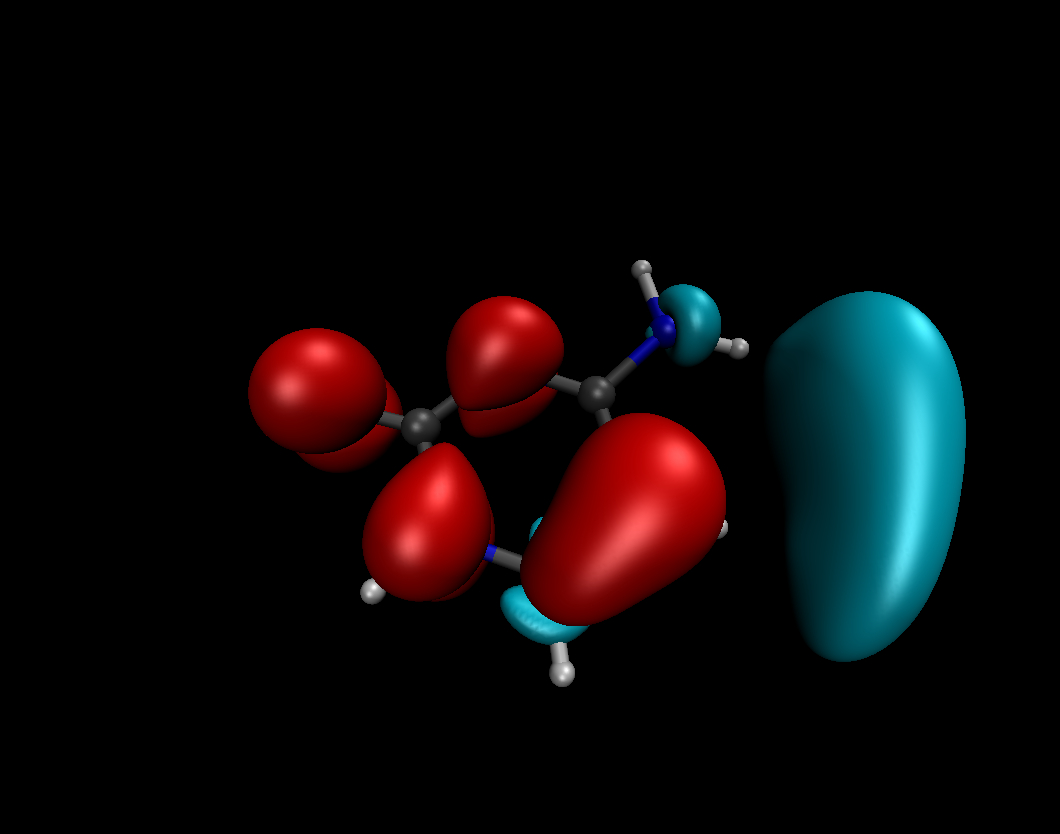
The Fukui functions and the Dual Descriptor
#The Fukui functions and the Dual Descriptor are suitable tools of the conceptual DFT designed to describe and predict the chemical reactivity. They describe how the electron density is modified after adding or removing some amount of electrons. They can predict where are the electrophilic/nucleophilic molecular sites. f-(r) indicates which are the preferred sites for an electrophilic attack while f+(r) indicates which are the preferred sites for a nucleophilic attack. Dual Descriptor combines the both properties of the Fukui functions.
See P. Geerlings, F. De Proft, and W. Langenaeker Chem. Rev. 103, 5, pp. 1793-1874 (2003)
DOCUMENTATION
Cite
New Insights in Quantum Chemical Topology Studies Using Numerical Grid-based Analyses. D. Kozlowski and J. Pilmé ,
J. Comput. Chem. 2011, 32, Issue 15, pp. 3207–3217
Promising insights in parallel grid-based algorithms for quantum chemical topology. H. Chevreau and J. Pilmé ,
J. Comput. Chem. 2023, DOI: 10.1002/jcc.27105
Recently cited by,
Anomalous p-backbonding in complexes between B(SiR3 ) 3 and N 2 : catalytic activation and breaking of scaling relations
Tore Brinck and Suman Kalyan Sahoo Phys. Chem. Chem. Phys., 2023, 25, pp. 21006
Understanding the Semiconducting-to-Metallic Transition in the CF2Si Monolayer under Shear Tensile Strain
Tarik Ouahrani and Reda M. Boufatah Crystals 2022, 12(10), pp. 1476
Relativistic Effects Stabilize Unusual Gold(II) Sulfate Structure via Aurophilic Interactions
Paul Jerabek, Archa Santhosh, and Peter Schwerdtfeger Inorg. Chem. 2022, 61, 33, pp. 13077–13084
LModeA-nano: A PyMOL Plugin for Calculating Bond Strength in Solids, Surfaces, and Molecules via Local Vibrational Mode Analysis
Yunwen Tao, Wenli Zou, Sadisha Nanayakkara, and Elfi Kraka J. Chem. Theory Comput. 2022, 18, 3, pp. 1821–1837
Halogen Bonding between Thiocarbonyl Compounds and 1,2- and 1,4-Diiodotetrafluorobenzenes
Lauri Happonen, J. Mikko Rautiainen, and Arto Valkonen Cryst. Growth Des. 2021, 21, 6, pp. 3409–3419
Template Synthesis of NPN′ Pincer-type Ligands at Titanium Using an Ambiphilic Phosphide Scaffold
Alex Moerman, E. Daiann Sosa Carrizo, Benjamin Théron, Hélène Cattey, Pierre Le Gendre, Paul Fleurat-Lessard, and Adrien T. Normand Inorg. Chem. 2022, 61, 19, pp. 7642–7653
Quantum chemical topology of the electron localization function in the field of attosecond electron dynamics
Angela Parise, Aurelio Alvarez-Ibarra, Xiaojing Wu, Xiaodong Zhao, Julien Pilmé, Aurélien de la Lande J. Phys. Chem. Lett. 2020, 9, pp. 844–850
Spin–orbit coupling as a probe to decipher halogen bonding
Jérôme Graton, Seyfeddine Rahali, Jean-Yves Le Questel, Gilles Montavon, Julien Pilmé, Nicolas Galland Phys. Chem. Chem. Phys. 2018, 20, 47, pp.29616–29624
Tutorials.
QTAIM ANALYSIS FROM WFN. CRITICAL POINTS AND POPULATIONS/CHARGES,
QTAIM ANALYSIS FROM WFN. LOCALIZATION/DELOCALIZATION INDICES,
ELF ANALYSIS FROM WFN,
QTAIM ANALYSIS FROM CUBE FILES,
ELF ANALYSIS FROM CUBE FILES,
QTAIM ANALYSIS FROM CUBE FILES WITH PERIODIC CONDITIONS,
Download Section
System requirements : Linux (GLIBC 2.4 or later), Intel/AMD (x86) processor(s), 64 bit
Licence agreement: Using TopChem2 requires agreement to the following conditions upon receipt of the program:
- I understand that copyright and intellectual property rights are retained by Julien Pilmé (Sorbonne University, Paris). TopChem2 is provided without any expressed or implied warranties of any kind.
- I agree to use this software for no profit purpose and I will not incorporate any part of the code in a commercial software. Neither TopChem2 nor any of its components may be sold, redistributed, repackaged, bundled, copied, modified, disassembled or reverse engineered in any form, either in whole or in part, without the explicit, written consent of Julien Pilmé.
- In no event shall the author be held liable for any damages of any kind arising directly or indirectly from the download, installation or use of TopChem2. Its compatibility with or effect on any computer hardware or software, or the reliability or accuracy of any results it produces.
Installation
A. Open a linux terminal and extract the files from the tar archive:
> tar -xvf TopChem2_cube64_X_XX.tar.gz
B. Change the directory to where the files are located:
> cd INSTALL_TOPCHEM64
C. Run the bash script installer, it does not require administrative privileges to run:
> bash installer.sh
D. Run >topchem2 h to display all available options
Versions
2025 (Latest v. 5.01) : Accept Licence Agreement and Download
Release
- Bugs that occurred when reading cube files with the input: command instead of wfn/wfx files have been fixed.
2025 (v. 5.00) : Accept Licence Agreement and Download

2025 (v. 4.00) : Accept Licence Agreement and Download
Release

2024 (v. 3.07) : Accept Licence Agreement and Download
Release
- New TopChem2 v. 3.07 is now available: faster, free, and feature-packed! Perform ELF, QTAIM, and NCI analyses, as well as compute conceptual DFT descriptors, for quantum systems ranging from small to large.
2024 (v. 3.06_02) : Accept Licence Agreement and Download
2024 (v. 3.06) : Accept Licence Agreement and Download
Release
- Some bugs fixed in the population section. Fixed checked conflicts between variables
2023 (v. 3.05) : Accept Licence Agreement and Download
Release
- 1D and 2D plots
- Automatic mode and some bugs fixed
2023 (v. 3.04_1) : Accept Licence Agreement and Download
Release
- Improved visualization for NCI 2d grids
- VMD visualization state file is now produced for the QTAIM analysis (visualization of critical points locations)
2023 (v. 3.04) : Accept Licence Agreement and Download
Release
- 2eDMs Muller's approximation for the Electron density and ELF when post-SCF correlated methods are used
- Improved visualization for NCI 3d-grids and 2d grids
- ELF saddle points & CVB index
2023 (v. 3.03) : Accept Licence Agreement and Download
Release
- Fixed issue for the calculation of populations from large 3d-grids
- NCI 3d-grids and 2d grids are produced
- VMD visualization state file is automatically produced as an output file for the Reduced Density Gradient mapped onto the second eigenvalue of the electron density. The color types will be assigned and stored in some separated cube files. A vmd visualization state file called, file_nci.vmd is also created. This can be directly read in VMD as a 'Load visualization file' using
vmd -e file_nci.vmd and gnuplot file_nci.gnu
2023 (v. 3.02_1) : Accept Licence Agreement and Download
2023 (v. 3.02) : Accept Licence Agreement and Download
Release
- Accelerated process for generate Electrostatic Potential 3d Grids and MESP analysis
- Molecular Electrostatic Potential Isosurfaces interfaced with vmd
2022 : Accept Licence Agreement and Download
Release
- WFN & WFX input file
- Accelerated process for generate three-dimensional grids and to assign grid point to basins.
- New Conceptual DFT section.
- ELF output files interfaced with vmd.
- Several basin analysis algorithms are now available.
- Fukui functions and Dual Descriptor grid are produced within FMO description using the Koopmans approximation and the Fukui weighted method.
- VMD visualization state file are automatically produced as an output file for the ELF and the Dual Descriptor. The color types will be assigned and stored in some separated cube files. A vmd visualization state file called, file_elf_ebas.vmd is also created. This can be directly read in VMD as a 'Load visualization file' using
vmd -e file_elf_ebas.vmd
2021 : Accept Licence Agreement and Download
Release
- Parallelized (OpenMP) fast process in order to generate three-dimensional grids and to assign grid point to basins.
- Voronoi assignment
To Do List
- User-defined scalar field
- Other formats (Molden)
- Localized orbital locator (LOL)
- Restrained ElectroStatic Potential (RESP) atomic charge
- Visualization QTAIM bond paths.
- Mathematica Input
- Graphic User Interface
CONTACT
Author. Julien Pilmé. Laboratoire de Chimie Théorique. Sorbonne Université
Email: julien.pilme@sorbonne-universite.fr