READ (LUCMD, *) DISPLC
Displacement taken in a numerical differentiation . This applies both
for a numerical molecular Hessian , as well as in
calculation of Raman intensities and optical activity . Read one more line specifying
value (*). Default is 10 ![]() a.u. However, note that this variable
do not determine the displacements used when evaluating numerical
gradient for use in first-order geometry optimizations with MP2 or CI
wave functions , which is controled by
the .DISPLA
keyword in the
*MINIMI
module.
a.u. However, note that this variable
do not determine the displacements used when evaluating numerical
gradient for use in first-order geometry optimizations with MP2 or CI
wave functions , which is controled by
the .DISPLA
keyword in the
*MINIMI
module.
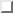 .
.
READ (LUCMD, *) NIPREAD (LUCMD, *) (IPART(IP), IP = 1, NIP)
Identify which fragments atoms belong to in
a dynamic walk. Read one more line specifying the number of
atoms (the total number of atoms in the molecule), then one more
line identifying which fragment an atom belongs to. The atoms in the
molecule are given a number, different for each fragment. See also the
discussion in Sec.  .
.
 .
.
READ (LUCMD, *) ANHFAC
Threshold for harmonic dominance. Read one more line specifying value. Default is 100. This is another way of changing the criterion for changes of the trust radius . See also the keyword .TRUST .
 .
.
READ (LUCMD,*) IWKIND
Desired Hessian index (strictly, of the totally symmetric block of the Hessian) at the optimized geometry. Read one more line specifying value. Default is 0 (minimum). Note that a stationary point with the wrong Hessian index will not be accepted as an optimized geometry.
READ (LUCMD, *) IRCSGN
Set the geometry walk to be an
Intrinsic Reaction Coordinate (IRC) as described in
Ref. [18, 19]. Read one
more line containing the sign (-1 or 1) of the reaction coordinate. It
cannot be decided in advance which reaction pathway a specific sign is
associated with. See also the discussion in Sec.  .
.
READ (LUCMD, *) NISREAD (LUCMD, *) (ISOTPS(IS), IS = 1, NIS)
Specify the isotopic constitution of the
molecule under investigation.
This is most interesting in dynamic walks , as well as
when using mass-scaled atomic coordinates . Note that the internal structure of
ABACUS uses Cartesian coordinates, and for vibrational analysis alone
the keyword .ISOTOP
in the *VIBANA
input section is to be
preferred. Note also that for defining center of mass
coordinates, there is a similar ISOTOP keyword in the general
input module, and this will become the only keyword for specifying
isotopic substitutions in later versions of the program. The
.ISOTOP
keyword in this input module (*WALK
) is to become
obsolete.
READ (LUCMD, *) XMXNUC
Maximum displacement allowed for any one atom as a result of the geometry update. Read one more line specifying value. Default is 0.5.
READ (LUCMD, *) TRUMX1
Set the maximum arc length in an Intrinsic Reaction Coordinate (IRC) walk . Read one more line containing the maximum arc length. Default is 0.10. Note that this arc length is also affected by the .TRUST keyword, and if both are specified, the arc length will be set to the minimum value of these to.
READ (LUCMD,*) IMODE
Mode to follow in level-shifted Newton optimizations for transition states . Read one more line specifying mode. Default is to follow the lowest mode (mode 1).
 .
.
READ (LUCMD, *) NSTMOM DO 265 IP = 1, NSTMOM READ (LUCMD, *) ISTMOM(IP), STRMOM(IP) 265 CONTINUE
Initial momentum for a dynamic walk .
Read one more line specifying
the number of modes to which there is added an initial momentum. Then
read one line for each of these modes, containing first the number of
the mode, and then the momentum. The default is to have no momentum.
See also the section describing how to perform a dynamic
walk, Sec.  .
.
READ (LUCMD, *) NZEROGREAD (LUCMD, *) (IZEROG(I), I = 1, NZEROG)
Set some gradient elements to zero. Read one more line specifying how many elements to zero, then one or more lines listing their sequence numbers.
 and
and  .
.
READ (LUCMD,*) IPRWLK
Set the print level in the prediction of new geometry steps. Read one
more line containing print level. Default value is the value of
IPRDEF in the general input module.
READ (LUCMD, *) RTMIN, RTRGOD, REJMIN, REJMAX
Limits on ratios between predicted and observed energy change. Read one more line specifying four values (*). These are respectively the bad prediction ratio, good prediction ratio, low rejection ratio and high rejection ratio. Defaults are 0.4, 0.8, 0.1, and 1.9.
READ (LUCMD, *) NREPSREAD (LUCMD, *) (IDOREP(I), I = 1, NREPS)
Consider perturbations of selected symmetries only. Read one more line specifying how many symmetries, then one line listing the desired symmetries. Note that only those symmetries previously defined to be true with the keyword .REPS from the ABACUS input modules will be calculated. This keyword thus represents a subset of the .REPS of the general input module.
READ (LUCMD, *) NUMNUC DO 7000 INUC = 1, NUMNUC READ (LUCMD, *) IATOM,(SCALCO(J,IATOM), J = 1, 3) 7000 CONTINUE
Scale the atomic coordinates. Read one more line specifying how many atoms to scale, then one line for each of these atoms (*) specifying the atom number and scale factors for all three Cartesian coordinates. Default is no scaling of the atomic coordinates.
DALTON.WLK file.
READ (LUCMD, *) TOLST
Threshold for convergence of the geometry optimization (on gradient
norm). Read one more line specifying the threshold (*). Default
is 10 ![]() .
.
READ (LUCMD, *) TRUSTR, TRUSTI, TRUSTD
Trust region information . Read one more line specifying three values (*): initial trust radius, factor by which radius can be incremented, and factor by which it can be decremented. Defaults are 0.5, 1.2 and 0.7, respectively; initial trust radius default is 0.3 if desired Hessian index is greater than zero. In dynamic walks the trust radius is by default put to 0.005, and in walks along an Intrinsic Reaction Coordinate (IRC) the default trust radius is 0.020. For dynamical walks the default increment and decrement factor is changed to 2.0 and 0.8 respectively.
READ (LUCMD, *) ZERGRD
Threshold below which gradient elements are
treated as zero. Read one more line specifying value (*). Default
is 10 ![]() . This keyword is mainly used for judging which modes are
symmetry breaking when using the keyword .KEEPSY
as well as
when deciding what step to take when starting a walk from a transition
state.
. This keyword is mainly used for judging which modes are
symmetry breaking when using the keyword .KEEPSY
as well as
when deciding what step to take when starting a walk from a transition
state.