



Next: Getting the wave function
Up: Getting started with DALTON
Previous: The MOLECULE input file
Contents
Index
The first calculation with DALTON
If we have made two input files, one corresponding to
DALTON.INP and one corresponding to the MOLECULE input file, we
are ready to do our first calculation. Examples of input files can
also be found in the dalton/test directory hidden inside the
test jobs. You can execute one of the individual test jobs
without the TEST script, for example:
> ./energy_nosymm
This will create four files, but right now we are just interested in
the .dal and the .mol files. In this particular example
they will be energy_nosymm.dal and
energy_nosymm.mol. Have a look at some of these input files to
get a head-on start on running different kinds of DALTON jobs.
Calculations with DALTON is most conveniently done using the
supplied shell script dalton. Thus,
to run a calculation of 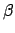 (first
hyperpolarizability)
with input available as
(first
hyperpolarizability)
with input available as beta.dal on the HCl molecule, and with
molecule input available as hcl.mol, you would type
> dalton beta hcl
assuming dalton is available in your path. When the job is
finished, the output is copied back as beta_hcl.out. In case that the
dalton- and molecule-input has the same name we may write
dalton energy_nosymm, and the corresponding output file will
be named energy_nosymm.out. In addition, the program will copy
back a file named beta_hcl.tar.gz. This file contains, in
tar'ed and gzip'ed form, a number of useful interface and
post-processing files needed for post-DALTON programs, or needed when
restarting calculations.
There are several options to this script, which can be viewed by
typing dalton -h or just dalton. These options include:
- -b directory
- Prepend this directory to the list of directories where the program
should look for basis sets. Needed in case you want to use local
modifications of a given basis. The job directory and the DALTON basis set library
will always be included in the basis set directory list (in that search order).
- -d
- Removes the contents of the scratch directory before a
calculation starts in order to avoid inconsistencies between files.
- -D
- Do not remove the content of the scratch directory. By
default the scratch directory will be deleted. However, in order to do
a restart you may want to keep all files in this directory, and you
then need to add the -D option when submitting the job.
- -e ext
- Change the extension to the output file from
.out
to .ext.
- -f
- Copy the gzipped-file containing a variety of useful
interface files from your home directory to the scratch directory
before a calculation starts. This is needed in order to be able to
restart DALTON calculations, or if you want to use converged response
vectors in a different response calculations for the same molecule.
- -M mb
- Change the default size of the scratch
memory space to
mb Mbytes.
- -N nodes
- Number of nodes to be used in a parallel MPI calculation
controlled by running mpirun (or using mpiexec).
- -lam file
- Call "lamboot file" and "wipe file" before and after mpirun.
- -o filename
- Redirect the output normally printed in the
DALTON.OUT file in the temporary directory
to the file ``filename'' in the WRKDIR
directory. On a computer system with distributed scratch disk but a
commonly mounted home directory, this allows you to follow the
calculation without having to log into the compute nodes.
- -t tmpdir
- Change the
TMPDIR (the scratch disc area) to
tmpdir from the default scratch directory determined at
install-time.
- -w wrkdir
- Change the working directory to ``wrkdir'', that is,
change the directory in which the program searches for input files
and places the
DALTON.OUT file to ``wrkdir''.
In most cases, the DALTON.OUT file will contain all the
information needed about a given calculations. However, in certain
cases, additional information may be wanted, and this is contained
in various sets of auxiliary files. These files are copied back in the
tar'ed and gzip'ed file. This file may include the following set of
different files:
- DALTON.BAS
- Contains a dump of a complete molecule input file.
This file take maximum
advantage of formatted input, yet differences may occur compared to
the basis sets obtained from the basis set library due to the
restricted number of digits available in the standard-format output.
- DALTON.CM
- An output file that contains the most essential
information needed for calculation of shielding polarizabilities
and magnetizability polarizabilities. Most easily used together
with the analyzing program
ODCPRG.f supplied in
the
tools directory.
- DALTON.HES
- If the keyword
.HESPUN has been specified in
the
*VIBANA input module, the molecular Hessian
will be written
to this file in a standard format, which may be used as a start Hessian
in an first-order geometry
optimization, or as input to a ROA or
VCD
analysis with different basis sets/level of correlation for the
intensity operators and the force field. See also the
FChk2HES.f program in the tools directory.
- DALTON.IRC
- Contains information obtained from an Intrinsic
Reaction Coordinate (IRC) calculation, as described in
Sec. 7.2.1.
- DALTON.MOL
- Contains the information needed by the
MOLPLT-program for visualizing the molecular geometry. The
MOLPLT-program is distributed with the GAMESS-US program
package.
- DALTON.MOPUN
- Contains the molecular-orbital coefficients
printed in a standard format allowing the transfer of molecular
orbitals coefficients from one computer to another.
- DALTON.NCA
- Contains the information needed by the
MOLPLT-program for visualizing normal coordinates.
- DALTON.ORB
- Contains information about basis set and
MO-coefficients so that MO density plots may be generated using the
PLTORB program that comes with the GAMESS-US
distribution. Currently not supported.
- DALTON.TRJ
- Contains trajectory information from a direct
dynamics calculation as described in
Sec. 7.2.2.
- DALTON.WLK
- Contains information from the walk-procedure, and is
needed when restarting a walk (e.g. a numerical differentiation).
- molden.inp
- Contains the input required for visualizing the
results of the calculation using the MOLDEN program
(http://http://www.cmbi.ru.nl/molden/molden.html). Please also note that Jmol (http://jmol.sourceforge.net)
also can visualize many aspects of a DALTON calculation, including an
intrisic reaction coordinate calculation. Jmol uses the DALTON output
file (DALTON.OUT) for it's visualization.
- RESULTS.RSP
- Contains a brief summary of the results obtained
form the response functions that have finished. The program may use
this information to skip response equations that have already been
solved (for instance if the calculation crashed for some reason
during the calculation of a set of cubic response functions).
- RSPVEC
- Contains the converged response equations. The program
may use this to avoid repeating linear response equations. Thus, one
may use the converged response vectors of a linear response equation
in the calculation of quadratic response function, and there may
then be no need to solve additional response equations.
- SIRIUS.IFC
- Interface file between the wave function part of the
program and the property modules. Contains all the information
required about the optimized molecular wave function.
- SIRIUS.RST
- Contains restart information needed in case one
needs to restart the wave function part of the program.




Next: Getting the wave function
Up: Getting started with DALTON
Previous: The MOLECULE input file
Contents
Index
Dalton Manual - Release 1.2.1
![]() (first
hyperpolarizability)
with input available as
(first
hyperpolarizability)
with input available as